|
Related Topics: |
|
|
|
Current News |
|
Chemistry A to Z |
|
About Internetchemistry |
New target for custom-tailored antibiotics discovered |
|
More and more bacterial stems are developing resistance to previously life-saving antibiotics. Physicians have been warning that fatality rates from infections could increase dramatically in the very near future. Researchers at the Technische Universität München (TUM) have now cast light on a metabolic step that appears in many aggressive microorganisms like tuberculosis or malaria pathogens and that may provide a promising target for a new class of antibiotics. The researchers presented the results of their work in the chemistry journal "Angewandte Chemie". Antibiotics hinder the production of essential compounds in microorganisms and can thus hold harmful pathogens in check. However, ever more bacterial stems are developing multiple antibiotic resistances, thereby rendering previously life-saving medications ineffective. That is why researchers around the world are desperately searching for new reaction steps that are vital to microorganisms but play no relevant role in humans. Professor Michael Groll, Dr. Jörg Eppinger and Dr. Tobias Gräwert, biochemists at the Technische Universität München, and their team of researchers have described in detail the structural basis for just such a reaction step. |
|
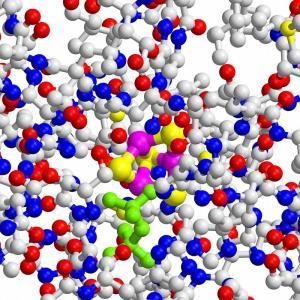
The cells of virtually all life forms synthesize essential natural substances belonging to the class of terpenes and steroids from the small isoprene building blocks dimethylallyl pyrophosphate (DMAPP) and isopentenyl pyrophosphate (IPP). Mammals and a large number of other organisms generate these essential metabolites via the so-called mevalonate pathway. But most human pathogens, including Plasmodium falciparum, have developed an alternate mechanism for producing these important substances. Now, this special pathway may spell doom for those bacteria. The TUM researchers have unraveled the structural basis of the terminal step in bacterial isoprene synthesis. The crucial enzyme has a most unusual structure, similar to a three-leaf clover, and may open a forceful line of attack for custom-tailored antibiotics. Research into the bacterial synthesis of isoprene building blocks was initiated as early as twelve years ago by Professor Adelbert Bacher in collaboration with Drs. Wolfgang Eisenreich and Felix Rohdich at the Chair for Organic Chemistry and Biochemistry. Over the years the team discovered most of the reaction steps of the new metabolic pathway. Yet the structure of the terminal step catalyzed by the IspH enzyme remained stubbornly elusive. Earlier measurements suggested that the active core must be an iron-sulfur cluster with three iron and four sulfur atoms. But other researchers questioned the results and for many years the crystal structure of the enzyme that would provide the proof could not be determined. The main problem was the oxygen sensitivity of the enzyme, which degenerates very quickly in air, thus losing both its structure and function. Only recently a group from the Justus-Liebig University in Gießen managed to determine the X-ray crystal structure of the enzyme’s open state. However, this structure provides hardly any information on the mutation process catalyzed by the enzyme. Professor Groll’s, Dr. Eppinger’s and Dr. Gräwert’s research team have now succeeded in cracking the closed state X-ray crystal structure that shows the precise folding pattern of the protein chain and the chemical environment of the active site cavity. The crystal structure opened the door to a detailed examination of the reaction mechanism using computer simulation and mutagen experiments in which Escherichia coli bacteria are coerced into synthesizing defective IspH enzymes. Thus, the X-ray structure, kinetic measurements and mutagenic analyses ultimately confirmed the unusual arrangement of three iron and four sulfur atoms in the central cavity, just as proposed years ago. “Now that the location, the chemical process and the involved helpers of the IspH reaction have been identified,” explains Groll, “we have a new angle of attack for developing substances that block the terminal step in the bacterial synthesis of isoprene building blocks, thereby killing pathogens in a very targeted way. Since the enzyme and the associated reaction do not appear in mammals these compounds should have few or no side effects in humans.” The research was funded by the Hans-Fischer-Gesellschaft and the Stifterverband für die Deutsche Wissenschaft (Project No. 11047: Forschungsdozentur Molekulare Katalyse). |
|
|
|